New project APVV 0009-10
Project of Slovak Research and Development Agency APVV 0009-10
Beta-lactamase genes of enterobacteria in the animal environment and bioaerosols (Duration May 2011 - October 2014)
The main object of the project is obtaining a new knowledge about development of betalactam resistance (penicillins and cephalosporins) in the relation to resistance mechanisms (ESBL TEM, ESBL SHV, blaCMY, CTX-M, KPC) in enterobacteria in animal environment (cattle and poultry) and bioaerosols (waste water stations).
Further intent of the project will be the study of development fluoroquinolone (PMQR) mechanisms of resistance as a most occuring resistance in slovak broiler farms. Molecular subtypisation of resistant strains will be oriented on rep-PCR, PFGE and Maldi toff mass spectra analysis as well as on the occurence of pandemic clone Escherichia coli ST131. Practical exploitation of results will be in veterinary diagnostics and therapy and in the education students and veterinary surgeons.
Methodology
We used veterinary version of Midivet kit for investigation of minimal inhibitory concentrations (Gattringer, Niks et al. 2002) from Bel-Miditech Bratislava (Table 1).
Table 1. Composition and extent of dilution series of antimicrobial agents in MidiVet kit for Escherichia coli including interpretation criteria (CLSI, 2008) for resistance assesment in terms minimum inhibitory concentration
| Antimicrobial agent | Range of dilution | Susceptible | Resistant |
| (µg/ml) | (µg/ml) | (µg/ml) | |
| Ampicillin (AMP) | 0.5–64 | ≤16 | ≥32 |
| Ampicillin+sulbactam(A+IB) | 0.5+0.2-64+32 | ≤16+8 | ≥32+16 |
| Ceftiofur (CFF) | 0.12-16 | ≤4 | ≥8 |
| Cefquinome (CFQ) | 0.25-32 | ≤16 | ≥32 |
| Ceftriaxon (CTR) | 0.5-64 | ≤32 | ≥64 |
| Ceftazidime (CAZ) | 0.2-32 | - | - |
| Gentamicin (GEN) | 0.25-32 | ≤4 | ≥8 |
| Kanamycin (KAN) | 0.25-32 | ≤16 | ≥32 |
| Streptomycin (STM) | 1-128 | ≤32 | ≥64 |
| Neomycin (NEO) | 0.5-64 | ≤16 | ≥32 |
| Nalidix acid (NAL) | 1-128 | ≤16 | ≥32 |
| Ciprofloxacin (CIP) | 0.03-4 | ≤2 | ≥4 |
| Enrofloxacin (ENR) | 0.12-16 | ≤1 | ≥2 |
| Chloramfenicol (CMP) | 0.25-32 | ≤16 | ≥32 |
| Florfenicol (FLO) | 0.25-32 | ≤8 | ≥16 |
| Tetracycline (TTC) | 0.25-32 | ≤8 | ≥16 |
| Cotrimoxazol (COT) | 0.1+1.9-3+61 | ≤3+61 | >3+61 |
There are two significant changes in breakpoints (CLSI 2013): Ceftriaxon ≥4mg /L and ceftazidime ≥16mg/L . There is ertapenem ( ≥2mg/L) instead of kanamycin in MidiVet kit.
Project phases
- VE01: Dynamics of antimicrobial resistance development in animal environment and bioarerosols (05/2011-12/2013)
- VE02: Genotypic determination of resistance mechanisms in enterobacteria (model E.coli). (10/2011-10/2014)
- VE03: The effect of zoohygienic parameters on the animal healthy status and the survival of antimicrobial resistant bacteria in the animal (poultry and cattle) environment, and animal waste-water station. (05/2011-05/2014)
- VE04: Creation of environmental E.coli antibiotic resistance mechanisms in slovak veterinary database. (12/2011-10/2014)
- VE05: The implementation of results in cattle farms and animal waste-water station. (06/2012- 10/2014)
Investigators
From Institute of Animal Physiology Slovak Academy of Sciences, Kosice:
Principal investigator:
Prof. DVM. Vladimir Kmet, DSc. - kmetv@saske.sk - http://home.saske.sk/~kmetv/
From University of Veterinary Medicine and Pharmacy, Kosice:
Responsible investigator
DVM. Jan Venglovsky, PhD. - jan@venglovsky.com - www.ramiran.net
Results in 2014
Biofilm production, virulence and betalactamase genes in environmental E. coli (Cornejova et al. 2014, symposium Biofilm 6)
| Strains with weak biofilm | Fylogenetic Group |
| CTX-M1, Int1,Tn3, iutA, luxS | 1xA (commensal) |
| CTX-M1, Int1, iutA, luxS | 1xA |
| CTX-M1, Int1, fimA, aer, iutA, luxS | 1xB2 |
| CTX-M1, Int1, fimA, afa, aer, iut A, luxS | 1xA |
| CTX-M1, iutA, luxS | 1xD (pathogen) |
| Cit, Int1, luxS | 1xA |
| IMP, Int1, luxS | 1xA |
| qnrS, luxS | 1xA |
| micrH47, luxS | 1xA |
| Strains with moderate biofilm | |
| CTX-M1, CTX-M2, Cit, qnrS, Int1, Tn3, iutA, fimA, kps, papC, ColE1, luxS | |
| fimA, aer, papC | 1xND |
| Strains with strong biofilm | |
| fimA, aer | 1xND |
| without genes | 1xND |
| Strains without production of biofilm | |
| CTX-M1,Cit, Int, iutA, luxS | 1xB2 (pathogen) |
| CTX-M1, Int1, Tn3, papC | 1xB2 |
| CTX-M1, Tn3, fimA, sfa, papC, micrH47 | 1xB2 |
| Int1, Tn3, fimA, luxS | 1xA |
MIC statistics of environmental (communal and animal waste water stations) Escherichia coli (2011-2014, n = 231), according CLSI 2013
| ATB | R | MIC XG | MIC 50 | MIC 90 |
| Ampicillin | 81.3 % | 54.1 | 128 | 128 |
| Ampicillin+sulbactam | 16.8 % | 8.0 | 8.0 | 32 |
| Ertapenem | 1.3% | 0,1 | 0.06 | 0.25 |
| Ceftiofur | 44.7 % | 1.9 | 4.0 | 32 |
| Cefquinome | 14.7 % | 1.8 | 1.0 | 32 |
| Ceftazidime | 3,4% | 0.8 | 0.25 | 8.0 |
| Ceftriaxon | 25.1% | 3.8 | 4.0 | 64 |
| Gentamicin | 13.4 % | 0.7 | 0.25 | 16.0 |
| Streptomycin | 58.4 % | 37.5 | 64 | 256 |
| Neomycin | 21.2 % | 6.8 | 4.0 | 256 |
| Spectinomycin | 17.7 % | 30.0 | 32 | 128 |
| Nalidix acid | 58 % | 29.6 | 64 | 256 |
| Enrofloxacin | 42.8 % | 1.5 | 1.0 | 32 |
| Ciprofloxacin | 38.1% | 0.6 | 0.25 | 8.0 |
| Tetracycline | 59.5 % | 9.6 | 16 | 32 |
| Chloramfenicol | 22,6 % | 8.0 | 8.0 | 64 |
| Florfenicol | 27.7 % | 9.5 | 8.0 | 32 |
| Cotrimoxazol | 54.5 % | 27.1 | 128 | 128 |
| Colistin | 0 % | 0.4 | 0.5 | 1.0 |
MIC statistics of animal environmental swabs and air (2011-2014, n=41) Escherichia coli , according CLSI 2013
| ATB | R | MIC XG | MIC 50 | MIC 90 |
| Ampicillin | 80.4 % | 58.8 | 128 | 128 |
| Ampicillin+sulbactam | 9.7 % | 7.2 | 8.0 | 16 |
| Ertapenem | 0% | 0,1 | 0.06 | 0.06 |
| Ceftiofur | 26.8 % | 0.9 | 0.5 | 16 |
| Cefquinome | 4.8 % | 0.7 | 0.25 | 8.0 |
| Ceftazidime | 0% | 0.4 | 0.25 | 2.0 |
| Ceftriaxon | 12,2% | 1.7 | 0.5 | 32 |
| Gentamicin | 9.7 % | 0.5 | 0.25 | 2.0 |
| Streptomycin | 48.7 % | 20.6 | 16 | 128 |
| Neomycin | 4.8 % | 3.6 | 4.0 | 8.0 |
| Spectinomycin | 14.6 % | 20.6 | 16 | 128 |
| Nalidix acid | 78 % | 37.3 | 64 | 128 |
| Enrofloxacin | 58.5 % | 2.1 | 2.0 | 16 |
| Ciprofloxacin | 48.7% | 1.1 | 2.0 | 8.0 |
| Tetracycline | 58.5 % | 8.1 | 16 | 32 |
| Chloramfenicol | 17 % | 6.9 | 4.0 | 64 |
| Florfenicol | 9.7 % | 7.5 | 8.0 | 8.0 |
| Cotrimoxazol | 24.3 % | 8.4 | 4.0 | 128 |
| Colistin | 0 % | 0.4 | 0.5 | 1.0 |
MIC statistics of Escherichia coli from poultry (2012-2014, n=153),
according CLSI 2013.
Partially published by Drugdova and Kmet, 2013
| ATB | R | MIC XG | MIC 50 | MIC 90 |
| Ampicillin | 84.3 % | 52.0 | 128 | 128 |
| Ampicillin+sulbactam | 8.5 % | 7.2 | 8.0 | 16 |
| Ertapenem | 1,31% | 0,1 | 0.06 | 0.06 |
| Ceftiofur | 41.1 % | 1.6 | 4.0 | 8.0 |
| Cefquinome | 15.6 % | 2.5 | 8.0 | 32 |
| Ceftazidime | 1,31% | 1.1 | 0.5 | 8.0 |
| Ceftriaxon | 7.1% | 3.0 | 4.0 | 16 |
| Gentamicin | 0 % | 0.3 | 0.25 | 0.5 |
| Streptomycin | 47.7 % | 20.3 | 16 | 256 |
| Neomycin | 16.3 % | 5.4 | 4.0 | 64 |
| Spectinomycin | 15 % | 19.6 | 16 | 256 |
| Nalidix acid | 86.2 % | 81.7 | 128 | 256 |
| Enrofloxacin | 75.8 % | 4.8 | 8.0 | 32 |
| Ciprofloxacin | 62.7% | 2.5 | 8.0 | 8.0 |
| Tetracycline | 67.9 % | 11.7 | 32 | 32 |
| Chloramfenicol | 16 % | 6.5 | 4.0 | 64 |
| Florfenicol | 26.1 % | 8.4 | 8.0 | 16.0 |
| Cotrimoxazol | 42.4 % | 17.2 | 8.0 | 128 |
| Colistin | 0 % | 0.3 | 0.25 | 0.5 |
MIC statistics of Escherichia coli from calves (2011-2014, n=163), according CLSI 2013.
| ATB | R | MIC XG | MIC 50 | MIC 90 |
| Ampicillin | 67.4 % | 31.7 | 64 | 128 |
| Ampicillin+sulbactam | 9.8 % | 5.9 | 4.0 | 16 |
| Ertapenem | 0% | 0,1 | 0.06 | 0.06 |
| Ceftiofur | 11 % | 0.4 | 0.25 | 8.0 |
| Cefquinome | 1.2 % | 0.5 | 0.25 | 8.0 |
| Ceftazidime | 0% | 0.4 | 0.25 | 4.0 |
| Ceftriaxon | 1.2% | 0.9 | 0.5 | 8.0 |
| Gentamicin | 6.1 % | 0.4 | 0.25 | 2.0 |
| Streptomycin | 69.3 % | 50.9 | 64 | 256 |
| Neomycin | 33.7 % | 9.8 | 4.0 | 128 |
| Spectinomycin | 27.6 % | 34.8 | 16 | 512 |
| Nalidix acid | 28.2 % | 9.1 | 4.0 | 256 |
| Enrofloxacin | 22 % | 0.5 | 0.13 | 16 |
| Ciprofloxacin | 19.6% | 0.2 | 0.06 | 8.0 |
| Tetracycline | 63.1 % | 11.3 | 16 | 32 |
| Chloramfenicol | 33.1 % | 9.9 | 8.0 | 64 |
| Florfenicol | 25.7 % | 8.6 | 8.0 | 16.0 |
| Cotrimoxazol | 33.7 % | 13.4 | 8.0 | 128 |
| Colistin | 0 % | 0.4 | 0.5 | 1.0 |
Results in 2013
1) Identification of resistance mechanisms in coagulase negative staphylococci of food and animal and environmental origin (Kmet, Strakova 2013).
The goal of this study was to investigate the presence of resistance mechanisms in coagulase negative staphylococci (CoNS) isolated from foods (nonpasteurised cow milk and ovine cheese) and animals (fecal swabs from poultry, pigs, calves and animal environment).
Results. The percentages of resistance detected in the food vs. animal (and environmnetal) CoNS isolates were as follows: AMP (4,2% vs. 11,1%), OXA (21,1% vs 39,7%), GEN (2,8% vs. 11,1%), CIP (0% vs. 36,5%), ERY (4,2% vs. 9,5%), CLI (2,8% vs. 12,7), VAN (5,6% vs.4,7%), TTC (7% vs. 42,8%), CMP (2,8% vs. 0%), TRI ( 22,5% vs. 50,8%) and COT (15,5% vs. 14,3%). All the studied CoNS were susceptible to cefalotin and linezolid. MRCoNS of food vs animal isolates were present in 12,7% vs. 34,9 %. MRCoNS phenotype corresponded with the presence of mecA gene. The percentage of penicillin resistance phenotype in food and animal isolates were 9,8% vs. 11,1%. Both group of strains contained blaZ gene, also. Genes tetK and msrA were detected more frequently while aac(6')-Ie-aph(2")-Ia, aph(3')-IIIa and aadA only in individual food strains. The low diversity of antibiotic resistance genes related to the lower percentage of resistance in food isolates. However one calf Staphylococcus haemolyticus strain contained blaZ , mecA, ermC, msrA, aac(6')-Ie-aph(2")-Ia, aph(3')-IIIa and tetK genes, what was the consequence of antibiotic therapy in the farm.
Conclusions. The prevalence antibiotic resistance in food CoNS was lower than in fecal animal and environmental isolates. Both group of strains contained mecA, blaZ, tetK and msrA genes. All staphylococci were susceptible to linezolid. This analysis suggests that food antibiotic resistant CoNS may be reservoir for human population.
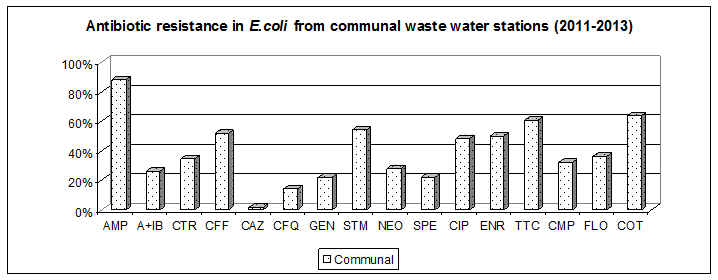
2) Antibiotic resistance in environmental Escherichia coli
Comparing the minimum inhibitory concentration of antibiotics (MIC90 ) in Escherichia coli from municipal waste water treatment plants against strains of animal wastewater treatment plant was found significantly higher MIC90 of cetriaxon 128mg / L vs. 4 mg / L.
In contrast, the MIC 90 level to fluoroquinolones (ciprofloxacin 8 mg / L, enrofloxacin 32 mg / L were similar for both groups of strains (municipal and animal). The strains consist of the genes CTXM-1, CMY-2 and plasmid quinolone resistance qnrS together with integron 1.
One strain of Escherichia coli B 17 isolated from biofilm stone belonged to pathogen group and contained CTX-M1, CTX-M2, CMY-2, integron1, transposon Tn3 and the virulence genes iutA and ColE1.
PCR method for detection of carbapenemases (blaNDM, blaVIM, blaIMP, blaKPC, blaOXA-like-48) was established. In one environmental isolate Escherichia coli, we demonstrated the presence of metallo-beta-lactamase IMP-type.
The results point to the fact that the genes of ESBL and metallo-beta-lactamase also circulate in the environment. It is probably the result of excessive use of antibiotics in the agglomeration Košice.
Results in 2012
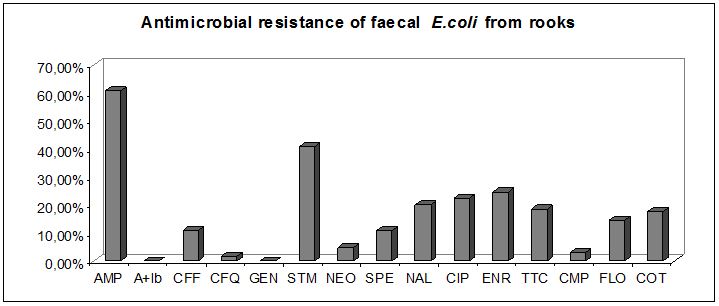
1) Escherichia coli in Rooks
In regards to antibiotic resistance studies in different model animals in the urban environment, we focused on the rook, which many behavioral and ecological aspects are important from an epidemiological point of view. This work investigates a total of 130 Escherichia coli strains isolated from rook faeces during two years period for antibiotic resistance and virulence.
The results showed that rooks can serve as reservoir of antibiotic resistant E. coli with avian pathogenic virulence factors for human population and potentially transmit such E.coli over long distances.
The betalactamase, quinolone and virulence genes in 25 E. coli strains from rooks (Kmet et al. 2013).
| Strains with CTX-M genes and virulence | Numbers of strains |
|---|---|
| CTX-M1, cvaC, iutA | 2 |
| CTX-M1, iutA | 1 |
| Strain with CTX-M, CMY-2 and virulence | |
| CTX-M1, CMY-2, papC,cva C, iutA | 1 |
| Strains with CMY-2 genes and virulence | |
| CMY-2, iss, ibeA, iutA, kpsII tsh | 3 |
| CMY-2, cvaC, iss, iutA, tsh | 1 |
| CMY-2, cvaC, iutA | 3 |
| CMY-2, cvaC, iss | 1 |
| CMY-2, cvaC | 1 |
| Virulence genes and integrase 1 | |
| cvaC, iss, iutA, tsh, Int1 | 2 |
| iss, iut, Int1 | 1 |
| iss, tsh, Int1 | 1 |
| iss, kps, Int1 | 1 |
| iss, Int1 | 2 |
| iut, Int1 | 1 |
| Strain with qnrS, integrase 1 and virulence | |
| qnrS, cvaC, iss, tsh, Int1 | 1 |
| Only virulence gene | |
| papC | 1 |
| iss | 1 |
| Strain with CMY-2 gene and integrase 1 | |
| CMY-2, Int1 | 1 |
| Total | 25 |
2) The comparison of virulence factors in environmental and uropathogenic Escherichia coli.
The aim of this study was PCR detection of virulence factors papC, iutA, iss, cvaC, kpsII, tsh (Delicato et al. 2003), ibeA (Germon et al. 2005), integron 1 (Mazel et al. 2000) and transposon 3 (Weill et al. 2004) in phenotype ESBL positive environmental (animal waste water) Escherichia coli with comparison of human uropathogenic Escherichia coli.
There was clear correlation between chu positivity (pathogen group B2), iutA and kpsII presence in the human urinal strains. However only one third of environmental strains belonged to the pathogen groups B2 and D. One pathogen (B2) environmental isolate E.coli ML 45 contained ibeA also with papC, iss , kpsII, integron 1, trasposon 3 and cit group genes (Perez-Perez et al. 2002). Maldi tof protein analysis revealed similarity of ML45 with human UPEC. Majority environmental strains containing integron 1, belonged to the A phylogenetic group of commensals with two or three virulence factors, however two strains were without virulence factors.
On the other hand occurence of ibeA gene was detected only in B2 pathogens. Results shows that some environmental Escherichia coli with virulence factors, mobile elements and ESBLs could be the source for the human population, also.
3) The comparison bacterial biodiversity Maldi tof and PFGE
Objective: To compare of Salmonella enterica serovar Typhimurium strains typing by PFGE and MALDI-TOF. Pulsed- field gel electrophoresis (PFGE) is widely used as an epidemiological tool for the typing, however is time consuming. In contrast, MALDI-TOF offers a rapid typing of bacterial strains immediately after bacterial identification.
Results: PFGE analysis differenciated all studied strains into 10 pulsotypes (X1, X1a, X2, X2a, X3, X4, X4a, X5, X6, X6a). The similarity of these PFGE profiles was analysed by MALDI Biotyper. A sufficient number of stable mass signals of major housekeeping proteins, mainly ribosomal proteins, can be used for estimation of the similarities between protein spectra of the same species. Distance level on the horizontal axis of MSP dendrogram indicates similarity of strains. There were two groups of pulsotypes: first consist of X1, X2, X4a and X5 pulsotypes (distance level around 450) and second group the remaining ones (distance level around 200). Moreover MALDI dendrogram recognised strain differences within the same pulsotype.
Conclusions: This study proves that MALDI MSP (main spectrum) dendrogram distinguished individually groups of Salmonella enterica serovar Typhimurium pulsotypes in our experiment. MALDI dendrogram recognized also strain differences within the same pulsotype.
Results in 2011
The main present and future risks for human population are resistant Escherichia coli with the production of ESBLs (extended spectrum betalactamases) including cefotaximases (CTX-M) and AmpC beta lactamases, carbapenemases (KPC) and plasmid quinolone resistance (PMQR).
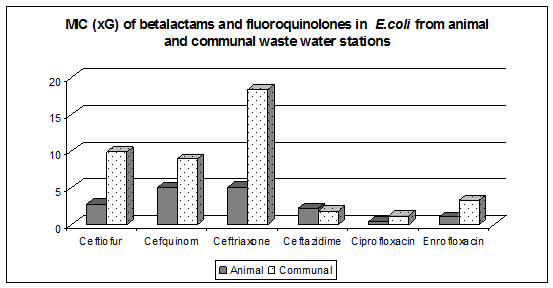
The purpose of the phase VE01 was the study of dynamics of antimicrobial resistance development in animal environment. MIC levels of betalactams were higher in Escherichia coli isolated from communal waste water stations in comparison with animal stations (Fig. 1) . Ertapenem resistant Escherichia .coli were not detected.
While working on a solution phase VE 03 “The survival of antimicrobial resistant bacteria in the animal environment” we determined total bacterial counts, total coliforms, moulds in aerosols and swabs taken at poultry abattoir during slaughtering in relation to the occurrence of extended spectrum betalactamases (ESBLs) in Escherichia coli.
The highest total bacterial counts (more than 106 CFU/m3) and counts of total coliforms (2x104 CFU/m3) were found at shackling, slaughtering and evisceration. In the course of dayshift the aerial contamination increased also in other stages, such as carving and packaging (total bacterial counts 103 CFU/m3 and total coliforms 101-103 CFU/m3, which can pose risk with regard to contamination of meat surface. Although the occurrence of ESBL was determined only quantitatively, it was recorded in both the clean and unclean sections of the operation.
Symposia in 2014
Brno, Czech Republic
Kmet V.: Development trends of antimicrobial resistance (invited lecture). In: 6 th Central European Veterinary Congress, 1.-2.4. 2014, Brno Exhibition Centre, Czech Republic
http://www.bvv.cz/en/cevc/profile-of-congress/Vienna, Austria
Cornejova T., Kmetova M., Kmet V.: Biofilm and ESBLs production in environmental Escherichia coli. In: Biofilms 6, International Conference on Microbial Biofilms, 11 - 13 May 2014, Vienna, Austria, Poster P-102, p. 117-118
http://biofilms6.univie.ac.at/program/Symposia in 2013
Berlin, Germany
V. Kmet, L. Majtanova, V. Majtan.: Typing human Salmonella enterica serovar Typhimurium isolates by pulsed field gel electrophoresis (PFGE) and matrix-assisted laser desorption/ionisation-time of flight (MALDI-TOF) mass spectrometry. ECCMID Berlin, 27-30 april 2013, Abstract R2712
http://www.escmid.org/escmid_library/online_lecture_library/Madrid, Spain
Kmet V., Strakova E.: Identification of resistance mechanisms in coagulase negative staphylococci of food and animal origin. Abstract 102, BioMicroWorld, 2-4 october 2013
FormatexSymposia in 2012
Copenhagen, Denmark
Kmet V., Faix S., Kmetova M.: The virulence factors in environmental Escherichia coli . Poster abstracts of 14 th International Symposium Microbial Ecology , Copenhagen 19-24 august 2012, Poster No. 378A, p.788-789
Valencia, Spain
KMEŤ, V , VENGLOVSKÝ, J.: The beta-lactam resistance in environmental Escherichia coli. Abstracts book 35th International Congress of the Society for Microbial Ecology and Disease (SOMED), 15-17 May 2012, Valencia, Spain, p. 84
http://www.somed2012valencia.com/img/ABSTRACTS_BOOK.pdf
Symposia in 2011
FEMS 2011, Geneve
http://www2.kenes.com/fems2011/sci/Documents/ScientificProgram.pdfhttp://www.saske.sk/atbres/pdf/Documents/Summary_FEMS_11.pdf
KMET, V., M. KMETOVA, Z. DRUGDOVA. Fluoroquinolone resistance of commensal Escherichia coli isolated from broilers. 4 th Congress European microbiologists FEMS 2011, Geneve, Poster. No.116, Abstracts in USB flash.
ISAH 2011, Vienna
http://www.isah2011.info/welcome.htmGREGOVA, G., VENGLOVSKY, J., KMET, V.: Microbial populations and antibiotic resistance in Escherichia coli isolated from poultry slaughterhouse. Proceedings of the XVth International Congress of the ISAH “Animal Hygiene and Sustainable Livestock Production”, Viena, 2011, Vol 3, pp. 1439-1442, ISBN 978-80-263-0012-0
VENGLOVSKY, J., GREGOVA, G., KMET, V., SASAKOVA, N.: Detection of airborne microorganisms and antibiotic resistance from animal housing facilities, Proceedings of the XVth International Congress of the ISAH “Animal Hygiene and Sustainable Livestock Production”, Viena, 2011, Vol 2, pp. 813-815, ISBN 978-80-263-0009-0
KMIN 2011, Plzen, CR (in Czech)
http://www.kmin2011.cz/pdf/program_kmin_verze-08.pdfhttp://www.saske.sk/atbres/img/Documents/Summary_Plzen_11.jpg
KMET V., KMETOVA M.: Beta-lactam and fluoroquinolone resistance of Escherichia coli isolated from poultry environment. Zborník abstraktov prednášok. Kongres klinické mikrobiologie a infekčních nemocí (KMIN) 2011, Plzeň, poster No. P10, s. 89, vyd. Euroverlag Plzeň, ISBN 978-80-7177-997-1
ZOONOSES 2011, Bratislava, SR (in Slovak)
http://www.sevs.sls.sk/kod04.htmKMET, V., KMETOVA, M., BUJNAKOVA, D., DRUGDOVA, Z. : Development of antimicrobial resistance in animal Escherichia coli in Slovakia. Zborník abstraktov, III. medzinárodná konferencia Zoonózy-spoločná ochrana zdravia ľudí a zdravia zvierat, Bratislava 2011, s. 23 , Vyd. MPRV SR,
ECOLOGY A VETERINARY MEDICINE, Kosice, SR (in Slovak)
http://www.uvm.sk/en/node/2930GREGOVA, G., VENGLOVSKY, J., KMET, V., SASAKOVÁ, N., ONDRASOVICOVÁ, O., VESZELITS LAKTICOVA, K.: Bioaerosols in animal housing facilities, Proceedings of VIIIth International Conference „ECOLOGY A VETERINARY MEDICINE“, 139-141, 2011, ISBN: 978-80-8077-249-9 (in Slovak)
References
- Gattringer R, M. Niks, R.. Ostertag, K. Schwarz, H. Medvedovic, W.Graninger, A. Georgopoulos, 2002: Evaluation of MIDITECH automated colorimetric MIC reading for antimicrobial susceptibility testing. J Antimicrob. Chemother. 49, 651-659
- Kmet V.: Host-microbial interplay in animal digestive tract and antimicrobial resistance in Escherichia coli (Scientific monograph in Slovak). Ed. Tribun EU Brno, 2009, 97 pp. ISBN 978-80-7399-554-6.
- Kmet V., Bujnakova D., Novak S.: Development of antibiotic resistance in Escherichia coli isolated from calves in Slovakia (in Slovak). Reporter of Bioveta SK, 2009, No. 6,
p. 6-7, ISSN 1337-6691 - Kmet V., Novak S., Miholics Š., Kmetova M. , Húska M.: Enrofloxacin resistance of faecal Escherichia coli in broilers and calves (in Slovak). Slovak Veterinary Journal, 35, No. 1, 2010, 42-43
- Drugdová Z., Kmet V., Bujnakova D.: Virulence factors in Escherichia coli isolated from chicken meat in Slovakia. J. Food and Nutr. Res. 49, 2010, 10-13
VUP - Kmet V., Kmetova M.: High level of quinolone resistance in Escherichia coli from healthy chicken broilers. Folia microbiologica 55, 2010, 79-82
SPRINGER - Kmet V., Piatnicová E.: Antibiotic resistance in commensal intestinal microflora. Folia microbiologica. Folia Microbiologica 55, 2010, 332- 335
SPRINGER - Gregova G., Kmetova M., Kmet V., Venglovsky J., Feher A.: Antibiotic resistance of Escherichia coli isolated from poultry slaughterhouse. Annals of Agricultural and Environmental Medicine 19, 75-77, 2012
AAEM - Kmeť V., Kmeťová M.: Detection of betalactamase genes in environmental Escherichia coli by DNA microarray (in Slovak). Slovak Veterinary Journal 37, No.1, 30-31, 2012, ISSN 1335-0099
- Kmet V., Drugdova Z., Kmetova M., Stanko M.: Virulence and antibiotic resistance of Escherichia coli isolated from rooks. Annals of Agricultural and Environmental Medicine, 20, 215-217, 2013
AAEM - Kmet V., Drugdova Z.: Antimicrobial susceptibility of microflora from ovine cheese. Folia microbiologica, 57, 291-293, 2012
Springer
Disclaimer & contact person
The information on this site is provided for educational and informational purposes only.
We don`t recommend or endorse any specific products that may be mentioned on the site. Reliance on any information provided by this site is solely at your own risk.
Dr. Vladimir Kmet
webpage: http://home.saske.sk/~kmetv/